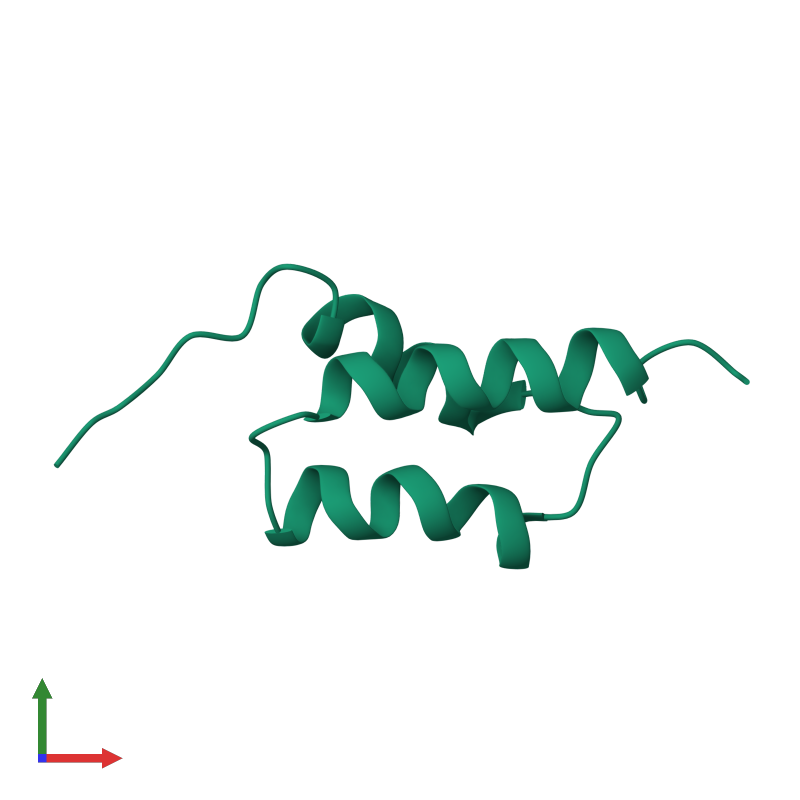
Entry STF0045
Protein A B-domain
Protein information
| Name of the protein: | Immunoglobulin G-binding protein A |
| Organism: | Staphylococcus aureus |
| Number of residues: | 58 |
| Related UniProt entry: | P38507 (Fragment: 212 - 270) |
| Related PDB entry: | 1BDD |
Visualize the data
Click here or on the image on the right to visualize the residues using JSmol. Warning: JSmol is known to load slowly on certain browsers, depending on the size of the macromolecule. The applet is optimized for Chrome, other browsers have limited support.
Experiment sets
STRONG
Method: Native exchange NMR
Conditions: pH 7.0; 20.0 Celsius; Probes: 28
Related publication:
PMID 9232646
Experiment details: "Equilibrium hydrogen exchange experiments were performed by dissolving lyophilized protein directly in potassium phosphate buffer (100 mM, pH* 7.0) in D2O containing various concentrations (0 - 3.0 M) of GdmC1. The GdmCl solution in D2O was prepared by addition of a measured volume of D2O to a weighed sample of deuterated GdmCl. Exchange rates were measured by recording 1H-15N HSQC spectra as a function of time without removing the sample from the magnet."
Protection threshold: ΔG(op) > 6 kcal/mol
Sequence:
TADNKFNKEQQNAFYEILHLPNLNEEQRNGFIQSLKDDPSQSANLLAEAKKLNDAQAPKA
CLICK TO DOWNLOAD SEQUENCE IN FASTA
STRONG residues
28: R;
35: L;
49: A;
50: K;
51: K;
CLICK TO DOWNLOAD LIST OF RESIDUES
EARLY
Method: Pulse labeling HDX NMR
Conditions: pH 5.0; 5.0 Celsius; Probes: 20
Related publication:
PMID 9232646
Experiment details: "Pulsed H/D exchange measurements were performed at 5 °C. The B-domain of protein A at a concentration of about 3 mg/mL was first unfolded in 6.0 M GdmC1 at pH 5.0 using 10 mM sodium acetate buffer in H2O. Folding was initiated by six times dilution with D2O buffer at pH* 5.0 and was allowed to continue for 6 ms before the labeling pulse was applied for 10 ms. The pulse buffer contained 0.5 M Tris adjusted to pH* 11.5 using KOD. This leads to a pulse of pH* 10.5 after mixing with the folding buffer. The exchange was quenched by D2O buffer containing 0.5 M Tris and 100 mM acetic acid at pH* 0.5. The final pH* of the solution was 4.8 after the quench."
Protection threshold: proton occupancy > 0.7 at 6 ms refolding time
Sequence:
TADNKFNKEQQNAFYEILHLPNLNEEQRNGFIQSLKDDPSQSANLLAEAKKLNDAQAPKA
CLICK TO DOWNLOAD SEQUENCE IN FASTA
EARLY residues
14: F;
15: Y;
16: E;
17: I;
18: L;
20: L;
22: N;
23: L;
28: R;
29: N;
31: F;
34: S;
35: L;
36: K;
47: A;
48: E;
49: A;
50: K;
51: K;
52: L;
CLICK TO DOWNLOAD LIST OF RESIDUES